Abstract
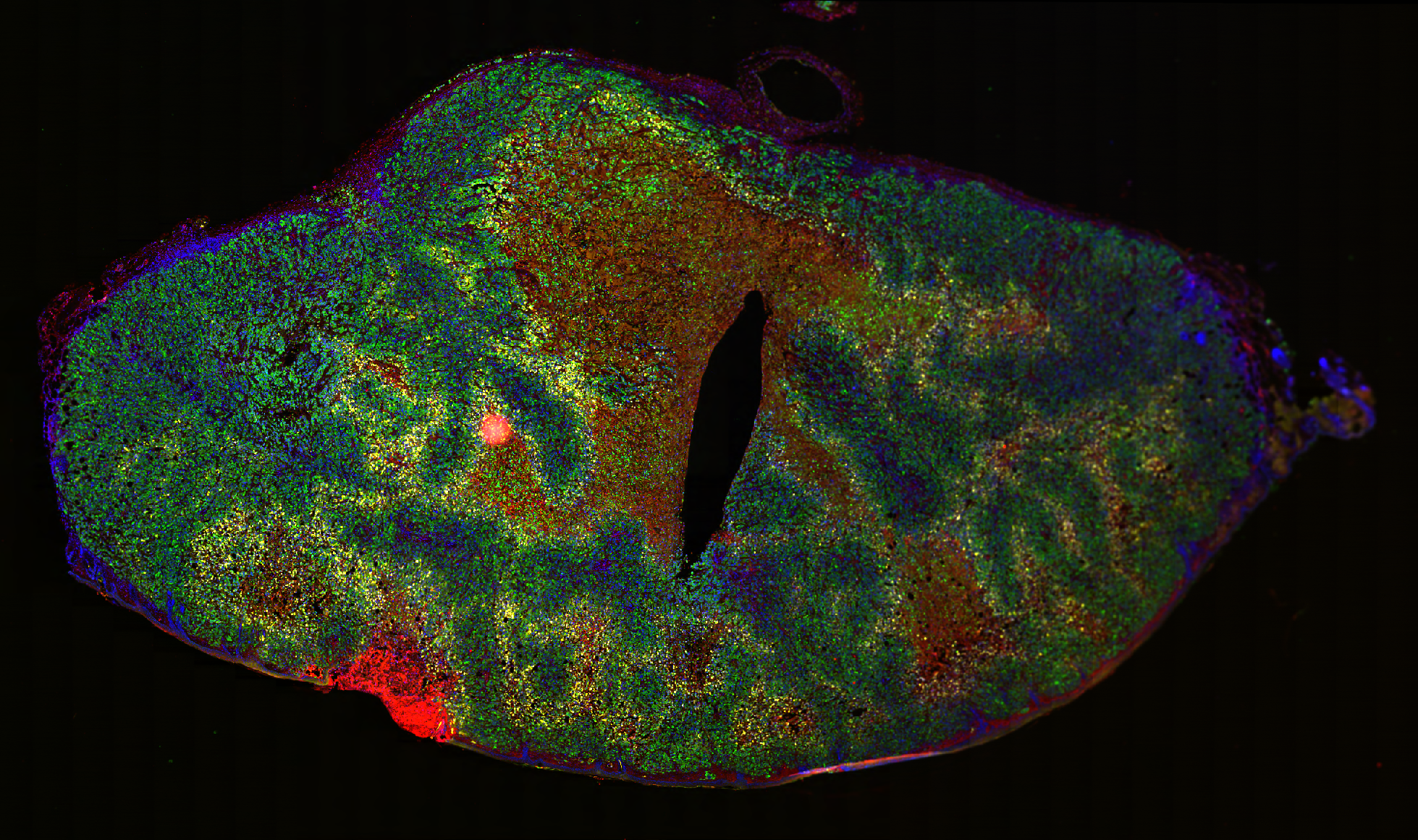
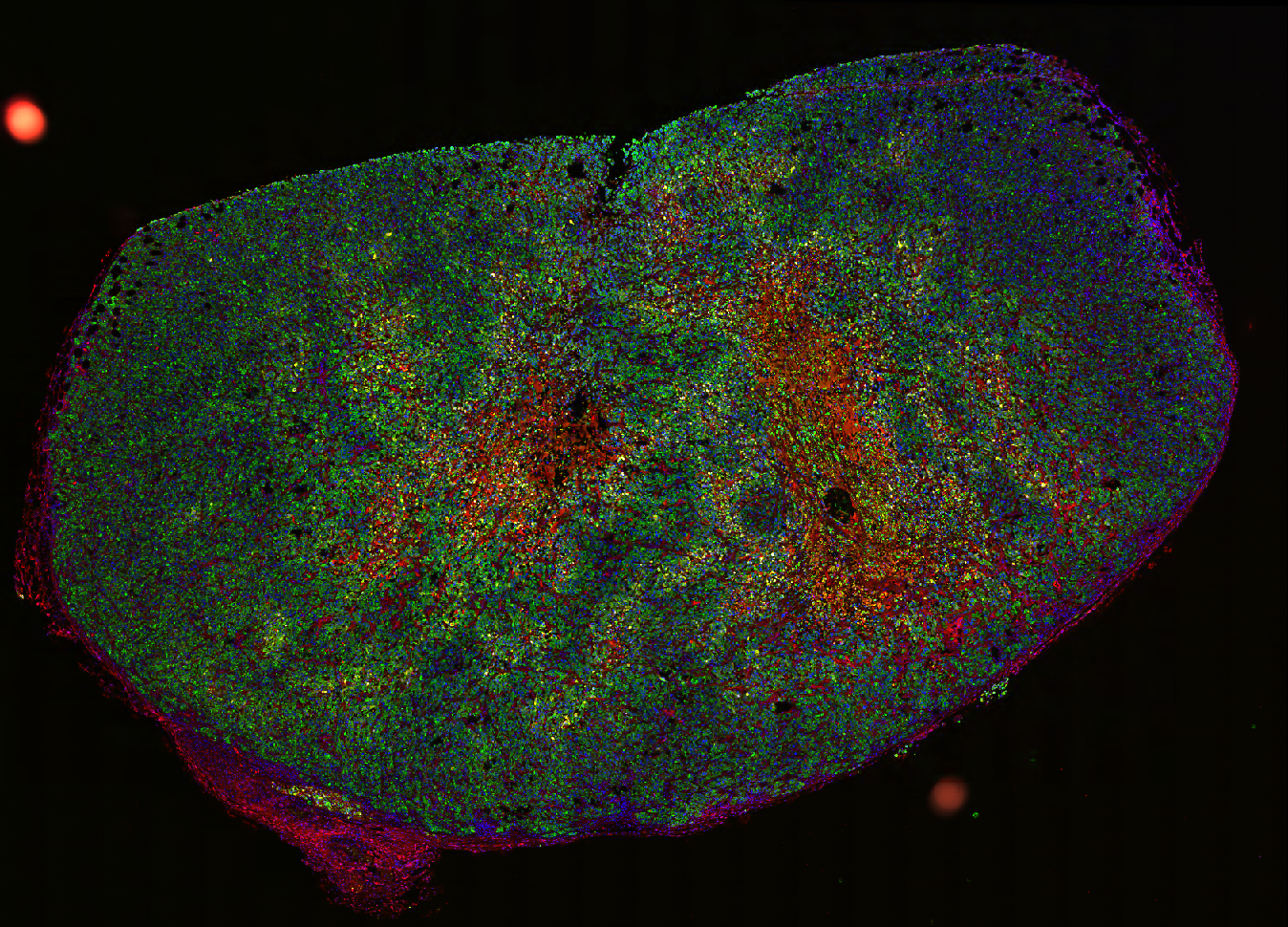
Dense, stroma-rich tumors with high extracellular matrix (ECM) content are highly resistant to chemotherapy, the standard treatment. Determining the spatial distribution of cell markers is crucial for characterizing the mechanisms of potential targets. However, an end-to-end computational pipeline has been lacking. Therefore, we developed a robust image analysis pipeline for quantifying the spatial distribution of fluorescently labeled cell markers relative to a modeled stromal border. This pipeline stitches together common models and software: StarDist for nuclei detection and boundary inference, QuPath’s Random Forest model for cell classification, pixel classifiers for stromal region annotation, signed distance calculation between cells and their nearest stromal border and Python for statistical modeling.