Abstract
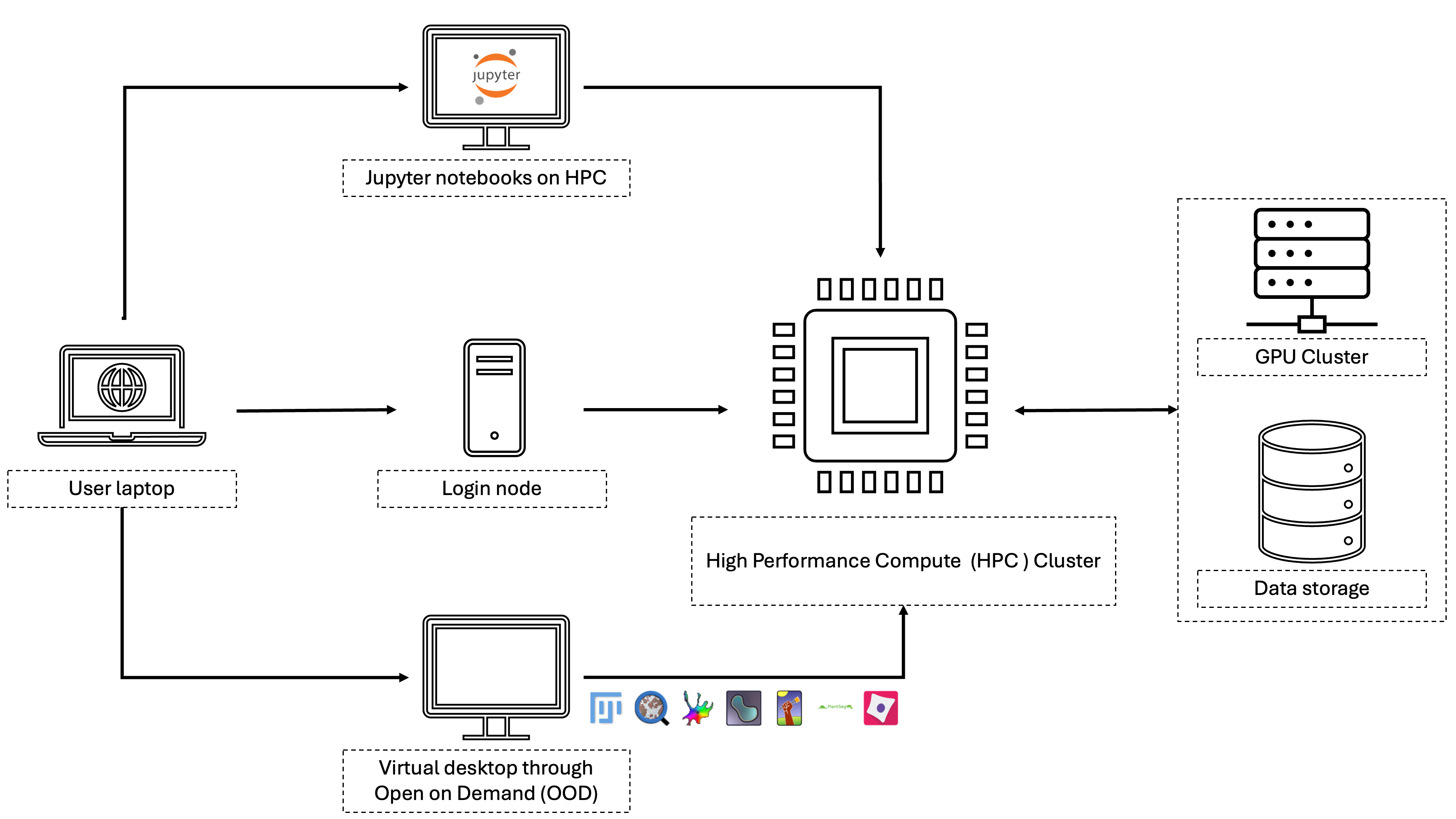
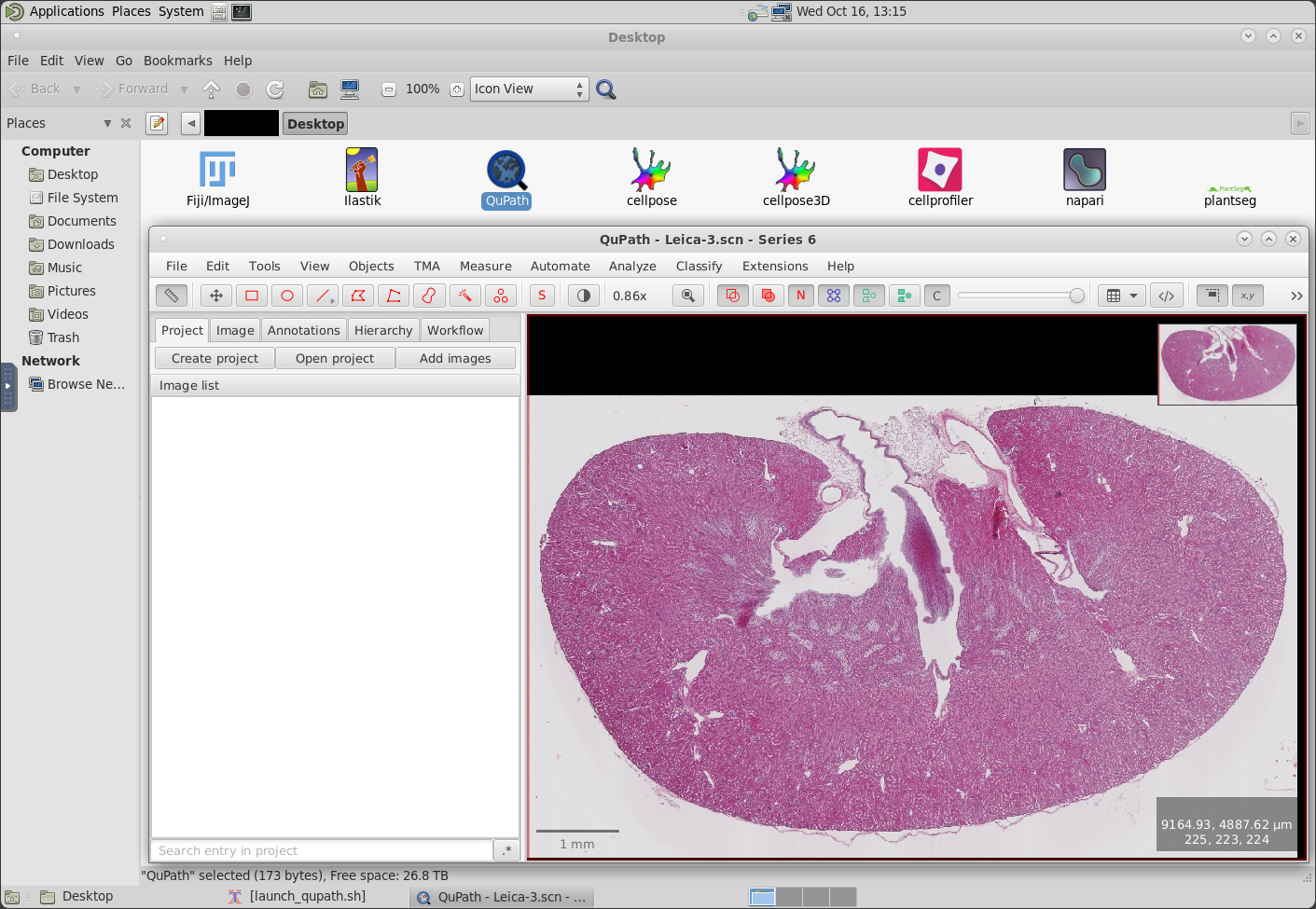
Quantitative analysis of microscopy data is crucial for scientific discovery. Although there are many open-source tools available for bioimage analysis, installing these tools can sometimes be complex. Additionally, researchers often encounter limitations when trying to use these tools on local resources. O2 offers good resources and easy access to data, but installing tools and accessing them remains a challenge. The aim of this project is to make bio-image analysis tools more accessible by deploying them on O2. The project is divided into two main tasks: installing bio-image analysis tools and deploying deep learning models. For the first task, we will develop installer files that users can run on their O2 accounts. These installers will handle all necessary environment and platform dependencies. For the second task, we will deploy popular deep learning networks on O2, enabling researchers to train them with their data and export results. We will also establish pipelines for data loading and evaluation. Our hope is that this will alleviate some of the barriers and accelerate scientific discovery.
Project Funding
This project is generously funded by the Core for Computational Biomedicine (CCB) and Dean's Innovation Award at Harvard Medical School.